
SeqScrub: a web tool for automatic cleaning and annotation of FASTA file headers for bioinformatic applications
Data consistency is necessary for effective bioinformatic analysis. SeqScrub is a web tool that parses and maintains consistent information about protein and DNA sequences in FASTA file format, checks if records are current, and adds taxonomic information by matching identifiers against entries in authoritative biological sequence databases.
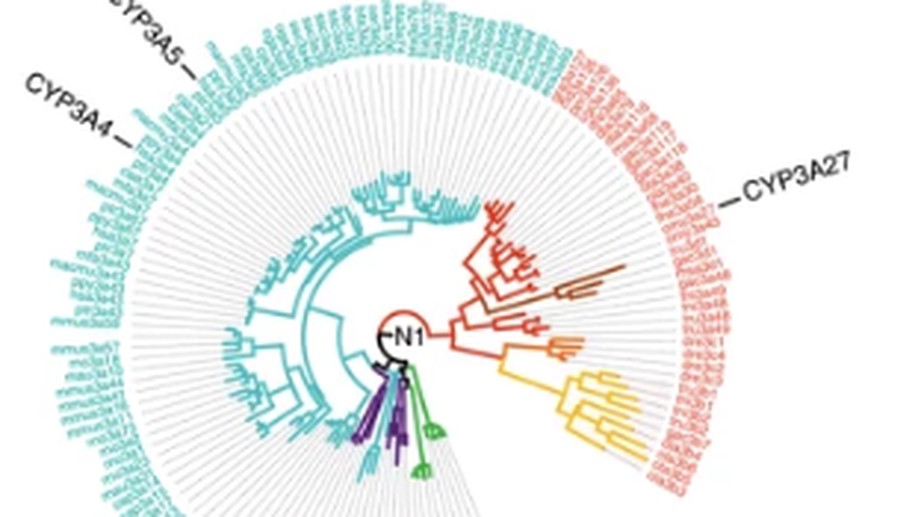
Engineering highly functional thermostable proteins using ancestral sequence reconstruction
Commercial biocatalysis requires robust enzymes that can withstand elevated temperatures and long incubations. Ancestral reconstruction has shown that pre-Cambrian enzymes were often much more thermostable than extant forms. Here, we resurrect ancestral enzymes that withstand ~30 °C higher temperatures and ≥100 times longer incubations than their extant forms.
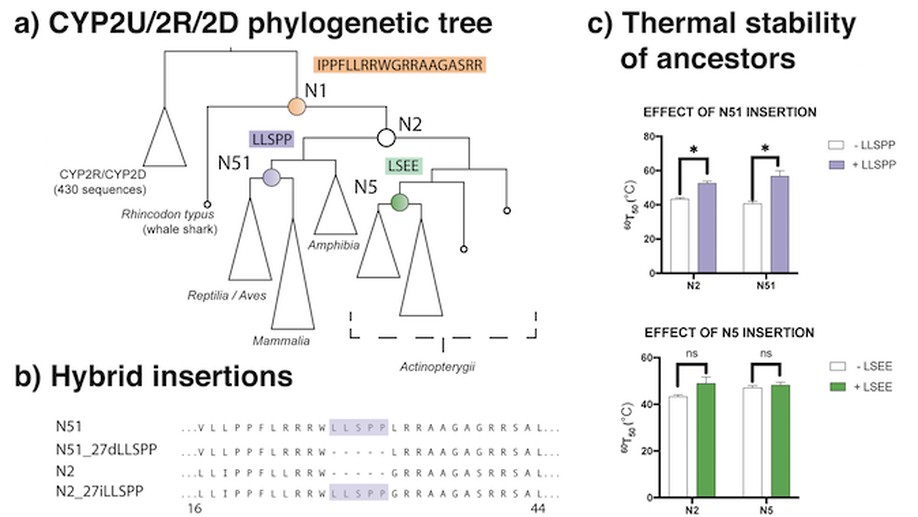
Engineering indel and substitution variants of diverse and ancient enzymes using Graphical Representation of Ancestral Sequence Predictions (GRASP)
Massive sequencing projects expose the extent of natural, genetic diversity. Here, we describe a method with capacity to perform ancestor sequence reconstruction from data sets in excess of 10,000 sequences, poised to recover ancestral diversity, including the evolutionary events that determine present-time biological function and structure.
We introduce a novel strategy for suggesting “insertion-deletion variants” that are distinct from, but can be explored alongside, substitution variants for creating ancestral libraries. We demonstrate how insertions and deletions can be used as building blocks to form “hybrid ancestors”; based on this strategy, we synthesise ancestor variants, with varying enzymatic activities, for wide-ranging applications in the biotechnology sector.